Multiple hypermutated SARS-CoV-2 sequences that may have been created by molnupiravir
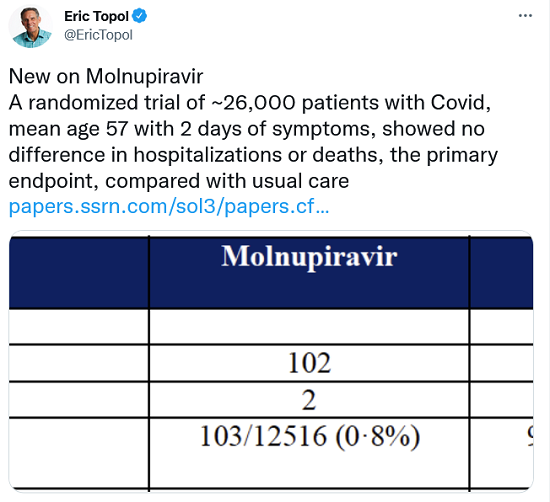
Molnupiravir, a drug used to treat SARS-CoV-2 infections, has received a lot of press attention since its launch, and not all of it has been positive.
Nearly one year ago, in a two part review of molnupiravir, William Haseltine at Forbes.com said “The FDA needs to tread very carefully with molnupiravir, the antiviral currently before them for approval. My misgivings are founded on two key concerns. The first is the drug’s potential mutagenicity, and the possibility that its use could lead to birth defects or cancerous tumors. The second is a danger that is far greater and potentially far deadlier: the drug’s potential to supercharge SARS-CoV-2 mutations and unleash a more virulent variant upon the world.”
Today, Ryan Hisner has posted a long thread about molnupiravir that we feel should be preserved publicly in case it is removed from Twitter. The thread looks at the approval process for the drug, then moves on to the potential for molnupiravir to cause hypermutated SARS-CoV-2 sequences, which is the part that we will focus on here.
It seems that, if this thread is correct, some of William Haseltine’s worst fears about this drug might be being realised.
Click on any of the images below to go to the complete Twitter thread.
… “Molnupiravir is more than just a crap drug. It works by causing random mutations throughout a virus’s genome. Potentially, this could accelerate the evolution of SARS-CoV-2 variants. See this @michaelzlin thread for more details on this aspect.”
But is there evidence molnupiravir is causing increased mutations, potentially fostering novel variants with unpredictable characteristics? AFAIK, no one is monitoring MOV use or even trying to determine its safety in this respect—regulatory negligence.
However, I’ve recently come across multiple hypermutated SARS-CoV-2 sequences that have every sign of having been created by molnupiravir. First, let’s review what some of these telltale signs are.
Molnupiravir randomly causes mutations throughout the viral genome. Random distribution of mutations is decidedly NOT what we see in the vast majority if highly mutated, chronic-infection sequences.
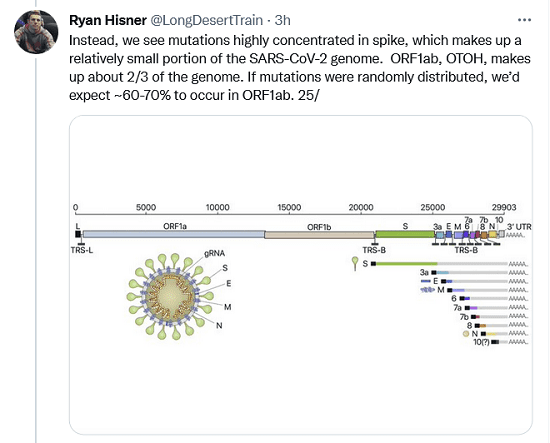
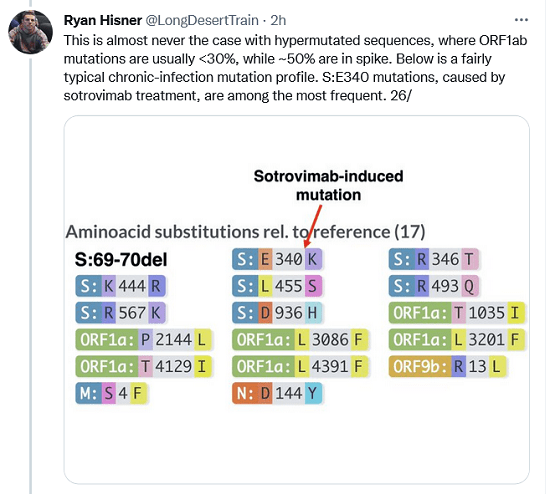
Instead, we see mutations highly concentrated in spike, which makes up a relatively small portion of the SARS-CoV-2 genome. ORF1ab, OTOH, makes up about 2/3 of the genome. If mutations were randomly distributed, we’d expect ~60-70% to occur in ORF1ab.
This is almost never the case with hypermutated sequences, where ORF1ab mutations are usually <30%, while ~50% are in spike. Below is a fairly typical chronic-infection mutation profile. S:E340 mutations, caused by sotrovimab treatment, are among the most frequent.
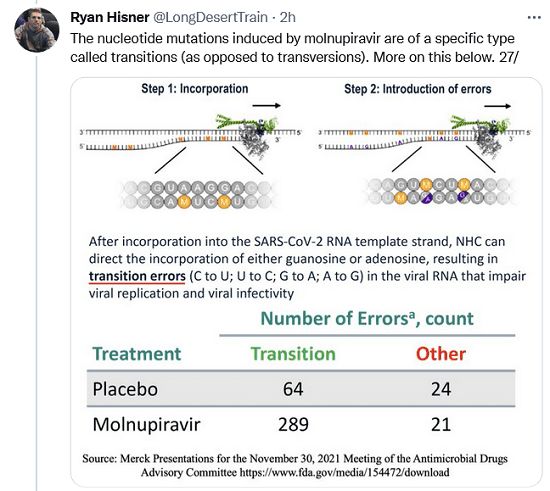
The nucleotide mutations induced by molnupiravir are of a specific type called transitions (as opposed to transversions). More on this below.
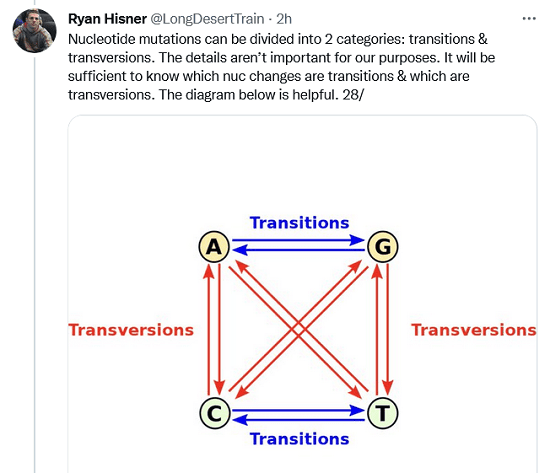
Nucleotide mutations can be divided into 2 categories: transitions & transversions. The details aren’t important for our purposes. It will be sufficient to know which nuc changes are transitions & which are transversions. The diagram below is helpful.
Transitions are T—>C, C—>T, A—>G, and G—>A. Everything else is a transversion: G—>T, T—>G, C—>G, G—>C, A—>C, C—>A, A—>T, and T—>A. (T = U for our purposes. Some diagrams/lists use T, some U.).
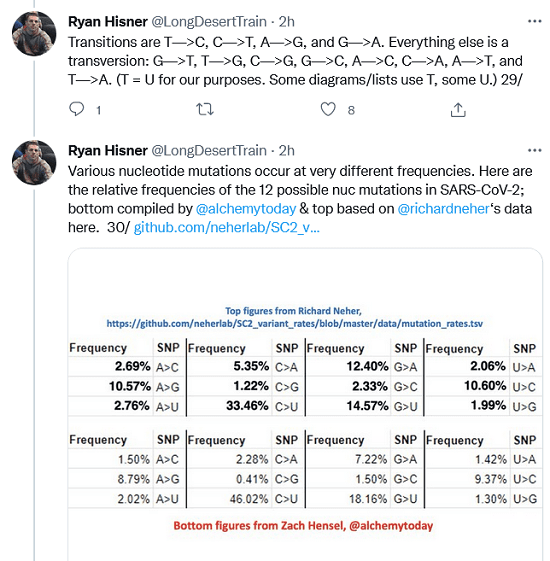
Various nucleotide mutations occur at very different frequencies. Here are the relative frequencies of the 12 possible nuc mutations in SARS-CoV-2; bottom compiled by @alchemytoday & top based on @richardneher‘s data here. https://github.com/neherlab/SC2_variant_rates/blob/master/data/mutation_rates.tsv
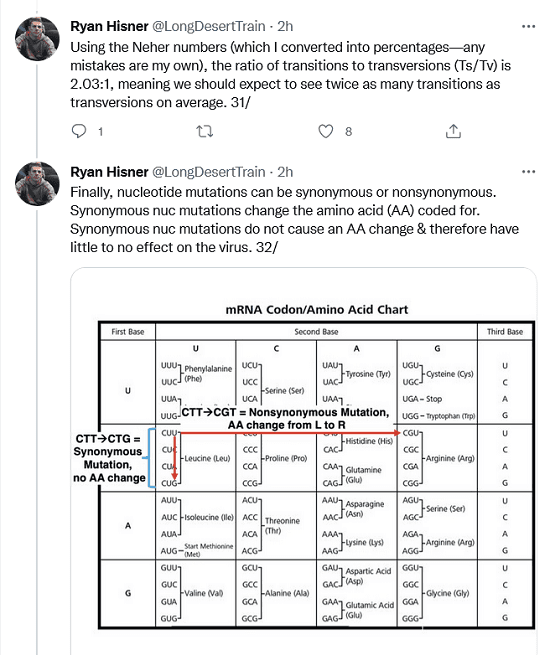
Using the Neher numbers (which I converted into percentages—any mistakes are my own), the ratio of transitions to transversions (Ts/Tv) is 2.03:1, meaning we should expect to see twice as many transitions as transversions on average. Finally, nucleotide mutations can be synonymous or nonsynonymous. Synonymous nuc mutations change the amino acid (AA) coded for. Synonymous nuc mutations do not cause an AA change & therefore have little to no effect on the virus.
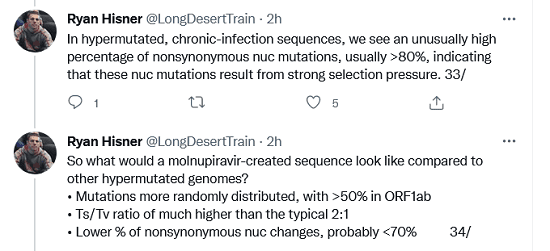
In hypermutated, chronic-infection sequences, we see an unusually high percentage of nonsynonymous nuc mutations, usually >80%, indicating that these nuc mutations result from strong selection pressure.
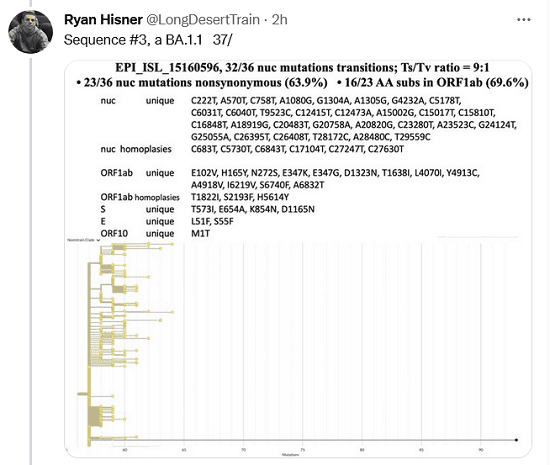
So what would a molnupiravir-created sequence look like compared to other hypermutated genomes? • Mutations more randomly distributed, with >50% in ORF1ab • Ts/Tv ratio of much higher than the typical 2:1 • Lower % of nonsynonymous nuc changes, probably <70%
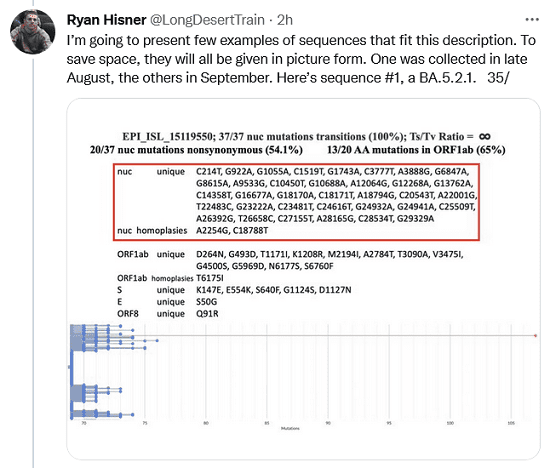
I’m going to present few examples of sequences that fit this description. To save space, they will all be given in picture form. One was collected in late August, the others in September. Here’s sequence #1, a BA.5.2.1.
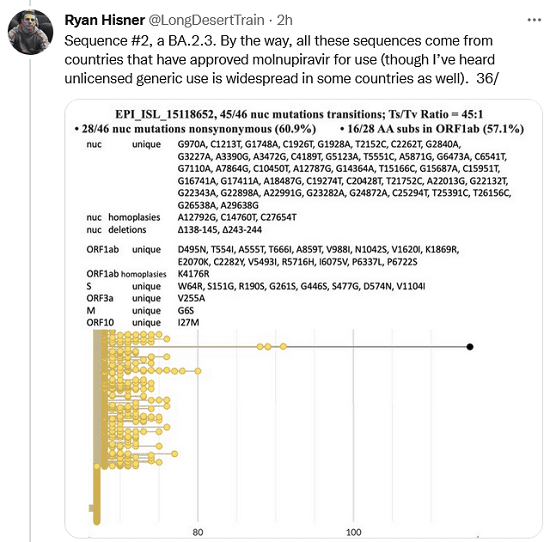
Sequence #2, a BA.2.3. By the way, all these sequences come from countries that have approved molnupiravir for use (though I’ve heard unlicensed generic use is widespread in some countries as well).
Sequence #3, a BA.1.1
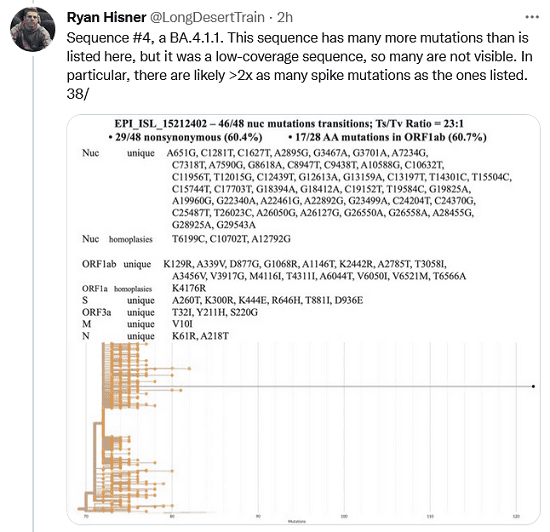
Sequence #4, a BA.4.1.1. This sequence has many more mutations than is listed here, but it was a low-coverage sequence, so many are not visible. In particular, there are likely >2x as many spike mutations as the ones listed.
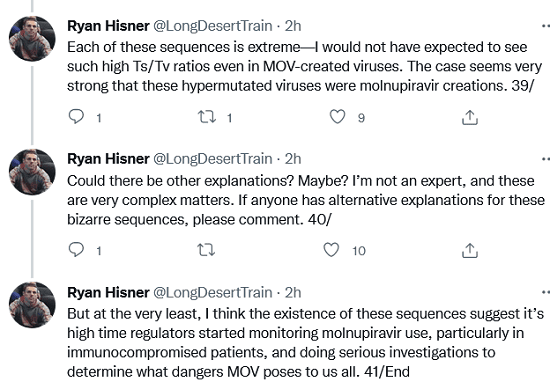
Each of these sequences is extreme—I would not have expected to see such high Ts/Tv ratios even in MOV-created viruses. The case seems very strong that these hypermutated viruses were molnupiravir creations.
Could there be other explanations? Maybe? I’m not an expert, and these are very complex matters. If anyone has alternative explanations for these bizarre sequences, please comment. But at the very least, I think the existence of these sequences suggest it’s high time regulators started monitoring molnupiravir use, particularly in immunocompromised patients, and doing serious investigations to determine what dangers MOV poses to us all.
Ryan Hisler – the entire Twitter thread on molnupiravir