Category: Evolution
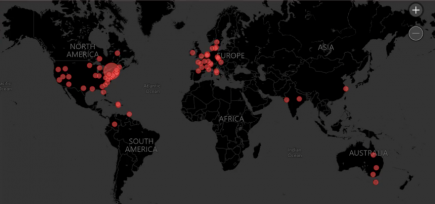
The stunning global growth of XBB.1.5
An incredible new visualisation showing the growth of XBB.1.5 across the globe, created by Mike Honey. More...
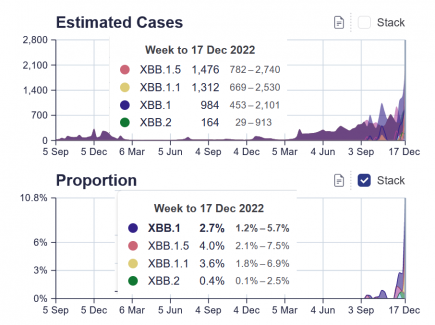
XBB.1.5 already outcompeting its closest relatives in England
The latest Sanger Institute data for England show that the recombinant Omicron subvariant XBB.1.5 is already outcompeting its familial lineages XBB, XBB.1.1 and XBB.2. More...
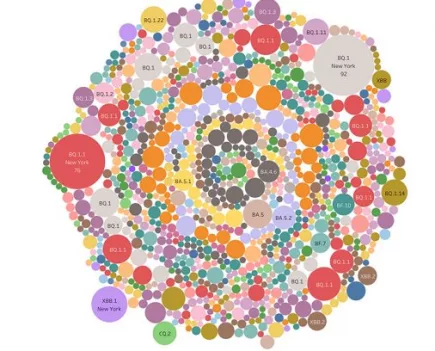
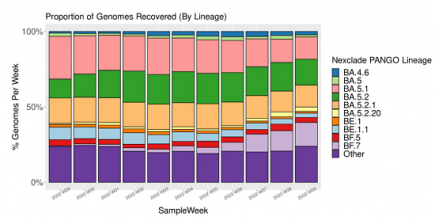
Denmark’s Covid variant soup is turning into a Covid variant smoothie
When we last checked in October 2022, Denmark had sequenced 120 different Covid variants – this month, they have sequenced 249 Covid variants. More...
Kenya: Beta-like SARS-CoV-2 variant found >1 yr after disappearance
A preprint out today gives details of a Beta-like SARS-CoV-2 variant with additional mutations recently found in Kenya. More...
Preprint: Neutralizing antibody escape of BQ.1, BQ.1.1, BA.4.6, BF.7 and BA.2.75.2
A recent preprint looks at the neutralizing antibody escape abilities for several new variants causing concern across the globe. More...
The world’s first XBB wave begins in Singapore
The world’s first wave of the recombinant SARS-CoV-2 variant XBB has apparently begun in Singapore. More...
Denmark: A soup of 120 Covid variants sequenced in one month
You may have heard the expression “a soup of Covid variants” recently.. More...
Multiple hypermutated SARS-CoV-2 sequences that may have been created by molnupiravir
Molnupiravir, a drug used to treat SARS-CoV-2 infections, has received a lot of press attention since its launch, and not all of it has been positive. More...
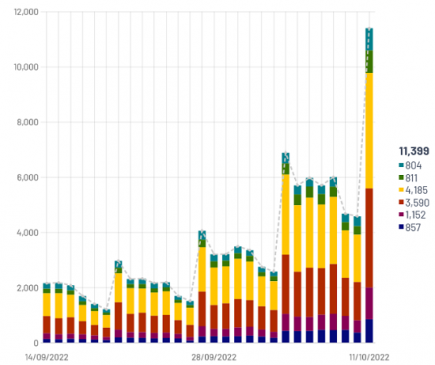
Growth advantage for BQ.1.1 is above 100% per week
The Omicron variant BQ.1.1 appears to be in a prime position to drive the coming winter wave in Europe. More...