Category: Origins
Bill Maher nails the Covid pandemic
‘A lot of the dissenting opinions that were suppressed and ridiculed at the time have proven to be correct.’ More...
Covid-19 Origins: The Raccoon Dog’s Breakfast *5 Updates*
The recently published scientific report about raccoon dog DNA being found in the Wuhan wet market has created headlines around the globe, but almost no one has read it, and even fewer seem to be aware of its conclusions. More...
WIV denied permission to use the BSL4 lab for SARS experiments
Damning information about the origins of Covid was released today – a 2018 cable from the US State Department reveals that the Wuhan Institute of Virology was refused permission to use its BSL4 lab for experiments on the SARS virus. More...
US Senate committee reports on the origins of the COVID-19 pandemic
“Substantial evidence suggests that the COVID-19 pandemic was the result of a research-related incident associated with a laboratory in Wuhan, China.” More...
Preprint: SARS-CoV-2 may have originated as an infectious clone assembled in vitro
A new preprint out today finds that SARS-COV-2 has “a synthetic fingerprint unlikely to have evolved from its close relatives.” More...
Research: “Omicron variant is highly likely a product of artificial genetic modification”
An extraordinary claim in a research paper just released: that the Omicron variant could be a product of artificial genetic modification. More...
Italy: SARS-CoV-2 found in patient samples from September 2019
“We conclude that a SARS-CoV-2 progenitor capable of producing a measles-like syndrome may have emerged in late June – late July 2019”. More...
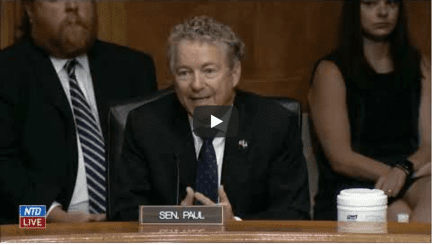
America finally starts asking the right questions about the origins of SARS-CoV-2
There were extraordinary scenes at a senate committee hearing last week as Senator Rand Paul forensically cross-examined scientists about the origins of the SARS-CoV-2 pandemic, and the gain-of-function experiments that may have caused it. More...
Preprint: Monkeypox has far more mutations than expected
“Researchers found the current Monkeypox strain diverges from the original strain by 50 single nucleotide polymorphisms (SNPs), and several mutations made the virus more transmissible.” More...