Category: Recombination
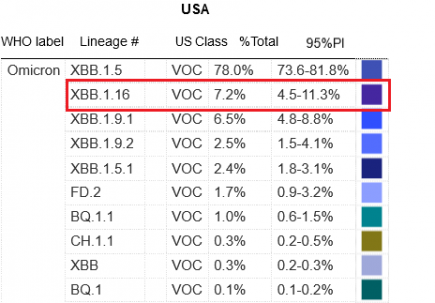
XBB.1.16 is the second most common variant in the US
XBB.1.16 has debuted on the CDC Covid variant list today in SECOND PLACE. More...
Deltacron recombinant XBC.1.6 at 30% of sequences in South Australia
We haven’t heard much about Deltacron recombinants recently, but one such variant is making real headway in South Australia – XBC.1.6 – is now at an astonishing 30% of SARS-CoV-2 sequences in that state. More...
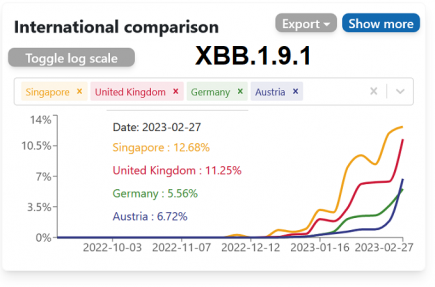
XBB.1.9.1 – a new variant of concern? – 4 updates
XBB.1.9.1, a new recombinant subvariant that is suddenly cropping up across the globe, is causing some concern amongst epidemiologists. More...
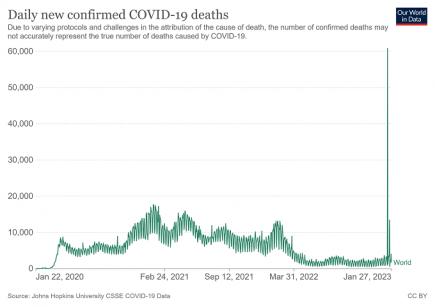
China: 60,000 Covid deaths finally added to global records
The 60,000 Covid deaths that were reported by China on the 14th January 2023 have finally been added to global Covid data records. More...
Sato Lab: Virological characteristics of XBB.1.5
Japan’s Sato Lab have announced new research into the Omicron XBB.1.5 ‘Kraken’ variant, confirming much about what is already known or suspected about its transmissibility. More...
Record Covid deaths in Japan, pneumonia increases in Italy
Just two days ago, the WHO warned of an increase in Covid deaths across the globe. More...
XBB.1.5 the most transmissible SARS-CoV-2 variant so far
Full video of the comments concerning XBB.1.5 at a World Health Organization briefing today. More...
XBB.1.5 at a staggering FORTY PERCENT of sequences in the US
These numbers were so shocking that, when they were leaked online yesterday, we didn’t believe them. More...
XBB.1.5 has a much higher hACE2 binding affinity than XBB.1
A new Twitter thread by Chinese researcher Yunlong Richard Cao about XBB.1.5 seems to confirm some of the anecdotal data collected recently about this hypertransmissable subvariant. More...
Recombination potential between SARS-CoV-2 and MERS-CoV
A June 2021 preprint looking at the potential for a MERS-CoV/SARS-CoV-2 recombination event is receiving a lot of social media attention today following rumours of MERS-CoV/SARS-CoV-2 infections at the World Cup in Qatar. More...