Category: Mutation
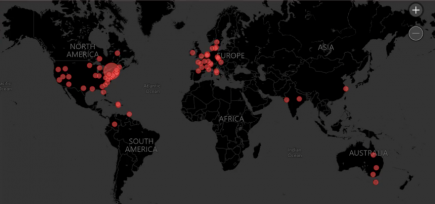
The stunning global growth of XBB.1.5
An incredible new visualisation showing the growth of XBB.1.5 across the globe, created by Mike Honey. More...
Bloom Lab: Why is XBB.1.5 so contagious?
One of the world’s top Covid labs explains how Omicron XBB.1.5 has learned new tricks to become more transmissible than other SARS-CoV-2 variants. More...
XBB.1.5 has a much higher hACE2 binding affinity than XBB.1
A new Twitter thread by Chinese researcher Yunlong Richard Cao about XBB.1.5 seems to confirm some of the anecdotal data collected recently about this hypertransmissable subvariant. More...
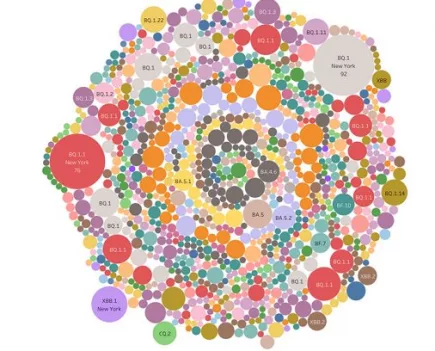
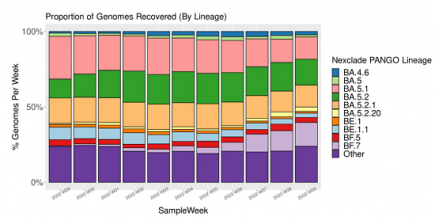
Denmark’s Covid variant soup is turning into a Covid variant smoothie
When we last checked in October 2022, Denmark had sequenced 120 different Covid variants – this month, they have sequenced 249 Covid variants. More...
Recombinant SARS-CoV-1 and SARS-CoV-2 variant found in Chinese bats
A new preprint out today has been looking at viruses in Chinese bats and has turned up some surprises – it looks like a recombinant variant of SARS-CoV-1 and SARS-CoV-2 has been found! More...
Canada: First evidence of deer-to-human transmission
A recent report from Sunnybrook Hospital in Toronto, Canada, gives details of a highly mutated strain of SARS-CoV-2, and evidence of deer-to-human transmission. More...
Preprint: Widespread wildlife exposure to SARS-CoV-2 in the Eastern United States
Species that tested PCR positive for SARS-CoV-2 included bobcats, opossums, squirrels, foxes, deer and rabbits. More...
Kenya: Beta-like SARS-CoV-2 variant found >1 yr after disappearance
A preprint out today gives details of a Beta-like SARS-CoV-2 variant with additional mutations recently found in Kenya. More...
Denmark: A soup of 120 Covid variants sequenced in one month
You may have heard the expression “a soup of Covid variants” recently.. More...
Multiple hypermutated SARS-CoV-2 sequences that may have been created by molnupiravir
Molnupiravir, a drug used to treat SARS-CoV-2 infections, has received a lot of press attention since its launch, and not all of it has been positive. More...