Tag: Variant of concern
UK: learning to live with COVID-19 cannot mean simply allowing infections to spread unchecked causing hospitalisations, illness – including Long COVID, and deaths
Joint Statement from the Association of Directors of Public Health and the Faculty of Public Health on the COVID-19 Roadmap. More...
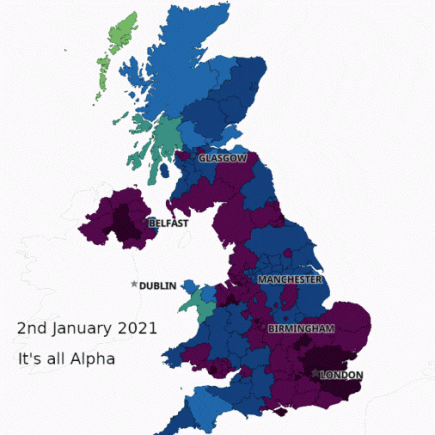
From Alpha to Delta – how Britain won and then lost the fight against coronavirus
After months of being ravaged by the Alpha variant (B.1.1.7) in 2020, Britain seemed to turn a corner in its fight with the coronavirus in early 2021. More...
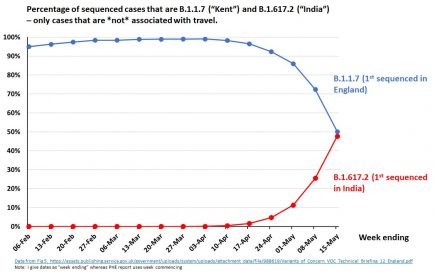
UK: PHE data shows B.1.617.2 now outcompetes B.1.1.7
More amazing work by Prof. Christina Pagel in deciphering the latest Public Health England data dumps for B16172. More...
WHO: Newly designated VOC within lineage B.1.617 – update on Delta coronavirus variant
In consultation with the WHO SARS-CoV-2 Virus Evolution Working Group, WHO has determined that viruses within the lineage B.1.617 have been characterized as a VOC. More...
WHO: B.1.617 Delta coronavirus strain is now a Variant of Concern or VOC
A World Health Organization official said Monday it is reclassifying the Indian Sars-Cov-2 variant B.1.617 as a “variant of concern,” indicating that it’s become a global health threat.
UK: surge testing for Delta coronavirus VOC B.1.617.2 begins in Bolton as some areas see 500% increase in cases
Mobile Covid-19 testing units are being rolled out in Bolton as surge testing gets underway in the borough’s coronavirus hotspots. More...
UK: B.1.617 found in 24% of coronavirus sequences in Bolton, mostly new Delta VOC B.1.617.2
Following a rise in cases in the UK and evidence of community transmission, PHE has reclassified B.1.617.2 as a Variant of Concern (VOC). More...
UK government poised to declare Delta coronavirus B.1.617 a Variant of Concern or VOC
The UK government is thought to have detected more than 40 clusters of the Indian Covid variant B.1.617 in the UK, and is on the verge of declaring it a “variant of concern”.
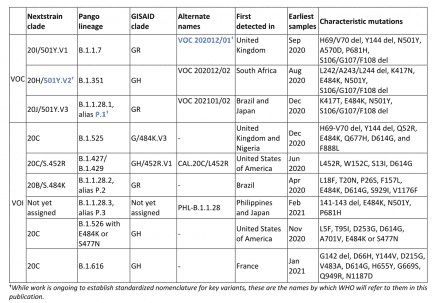
B1427/B1429 are listed as Variants of Concern by the CDC, as Variants of Interest (VOI) by WHO
The World Health Organization lists as B.1.427 and B.1.429 as Variants of Interest. More...
Sri Lanka: new SGTF coronavirus variant appears to have completely displaced B.1.411 within 10 days
Prof. Malavige said that currently, over 95% of the samples from Colombo, Kurunegala and Kalutara give an ‘S drop’, suggesting that these infections are due to this variant [B.1.1.7]. More...